Patented Workflow

Soil / Seabed Samples
- Our patented technology – developed to analyze surface soil or seabed samples (few mm3’s, from approx. 1 ft. depth) – recognizes otherwise undetectable hydrocarbon microseepage from prospective areas.

Field crew travel to the location to sample the area of interest, either in a grid pattern (covering a lease or area of interest) or visiting different target locations. Vast areas can be covered relatively quickly this way.
The samples are taken with specialized equipment and are stored in hermetic (completely airtight) conditions to avoid contamination.

DNA Analysis
- After the sample has been taken, first microbial DNA is extracted from this sample, producing tagged 16S rDNA data that is translated into species.
- The result of this analysis is hundreds of thousands (partly field type specific) biomarkers. The ‘DNA fingerprint’ of the soil sample.
- Taking a closer look at the complex composition of microbes, we find not only those that flourish from microseepage, but also those that perish by it—the latter have lower concentrations above sweet spots. More than 340,000 different species of microbes can be found in soil, of which a small number (from 50 to 200) react to these microseeps of gas.
- Biodentify creates a prospectivity ‘indicator’ on the new sampled locations using the DNA fingerprints, which is a value between -1 and 1. These values are plotted on a map which shows the prospect areas very distinctively and along one cross-section.
Schematic representation of the bacterial diversity and relative abundance within samples. Each colored bar represents a bacterial genus or family.
One soil sample could contain up to 35 million DNA signatures and ± 950,000 species. The extraction of microsomal chromosomal DNA is performed by mechanical disruption of the microbes using small beads and vigorous shaking.

Machine Learning
- Our database – with over 5800 samples from both onshore and offshore locations, the detailed DNA fingerprints, with related production data on biomarkers – is subsequently used as modeling input to our proprietary localized triple loop © computational model.
- Our trained model is capable of predicting the presence of a highly productive zone within a heterogeneous shale play, with an accuracy of >70% prior to drilling. For conventional plays, both on- and offshore, it is able to predict whether a prospect or well target location shows signs of hydrocarbons in the subsurface.
- Our model uses the difference in DNA fingerprints that are connected to highly productive areas and those correlated to unproductive (or dry) areas or locations.

There is too much data present in the sample for us humans to make sense of it alone, so our machine learning model finds the ‘signal’ in the ‘noise’. Only when enough biomarkers (50-200) are combined, a sufficiently accurate estimate can be produced on the relative prospectivity. The algorithms use machine learning to find a small difference in the DNA fingerprints related to microseepage and they neglect the influences of the climate, local spills or other external influences.

Prospect Map Delivered
- 8 shale plays in the US have now been extensively sampled, with different characteristics on productivity, the age of the producing interval, type of play (oil vs gas), geology and climate and soil type: Bone Spring (Permian), Bakken, Antrim, Avalon, Lewis, Haynesville, and Marcellus. In addition, studies have been carried out both on- and offshore the Netherlands as well as offshore Norway in the North Sea. Our recent 2019 pilot projects accurately predicted samples in conventional onshore Argentina, and in 2 shale plays in the USA.
- Final deliverable in all these projects: a detailed prospectivity map, highlighting highly productive zone (in red) versus low producing zone (blue) – prior to drilling, based only on shallow soil samples.
- And for our conventional fields we’ve delivered delineation surveys and prospectivity maps proving that we can identify charged reservoirs versus dry areas.
- Predictions can be made with an accuracy between 72-98% (based on the US Onshore benchmark study and pilot projects). Our data indicates that models and results are exportable between conventional and unconventional fields, and even continents. You can read more on our projects page.
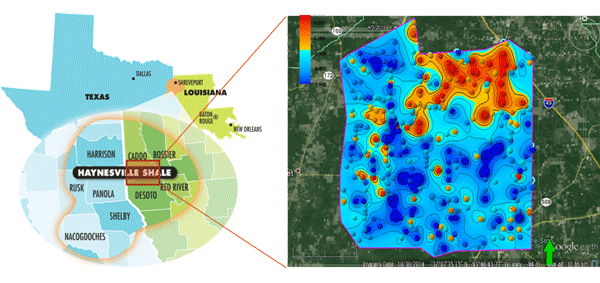
- Final deliverable in all these projects: a detailed prospectivity map.



